Plot a lot with ggplot2 to find plots
R-Ladies Colombo Meetup
Priyanga Dilini Talagala
2021/01/27
Tidy Workflow
Tidy Workflow
Tidy Workflow
The Datasaurus Dozen
library(datasauRus)library(ggplot2)library(gganimate)ggplot(datasaurus_dozen, aes(x=x, y=y))+ geom_point()+ theme_minimal() + transition_states(dataset, 3, 1) + theme(aspect.ratio = 1)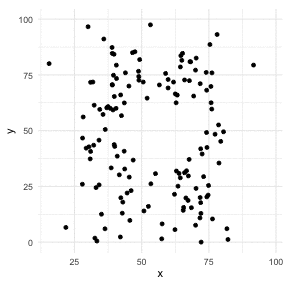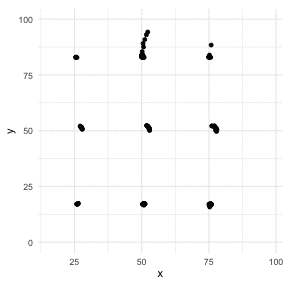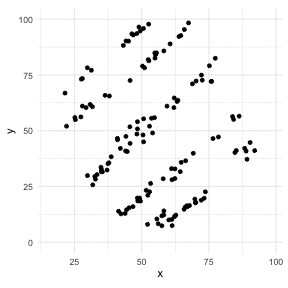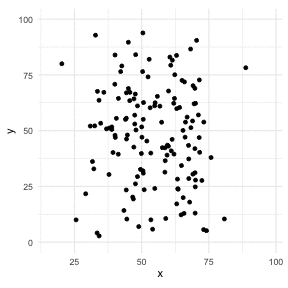
The Datasaurus Dozen
library(datasauRus)library(ggplot2)library(gganimate)ggplot(datasaurus_dozen, aes(x=x, y=y))+ geom_point()+ theme_minimal() + transition_states(dataset, 3, 1) + theme(aspect.ratio = 1)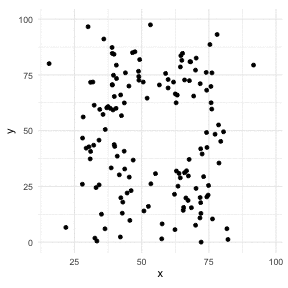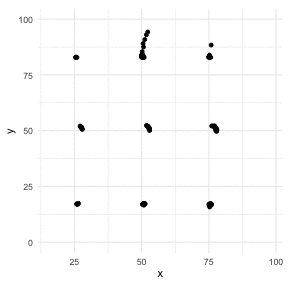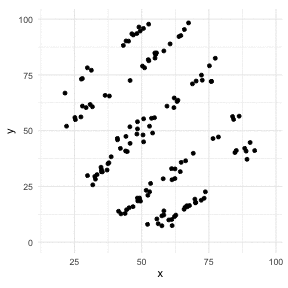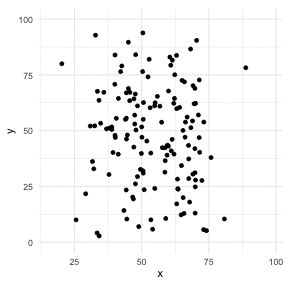
| Summary statistics | |
|---|---|
| X Mean | 54.26 |
| Y Mean | 47.83 |
| X SD | 16.77 |
| Y SD | 26.94 |
| Corr. | -0.06 |
The Grammar of Graphics
The Book
The Grammar of Graphics

R Base Graphics
The Grammar of Graphics
Pie Chart
Line Chart
Bar Chart
Scatterplot
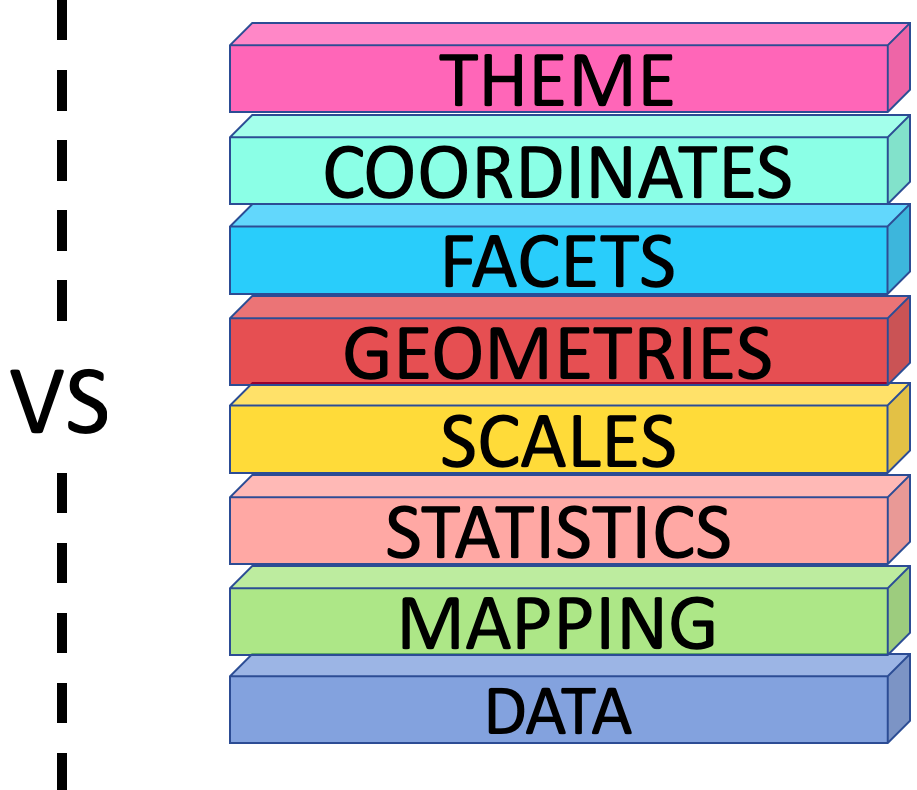
The Grammar of Graphics
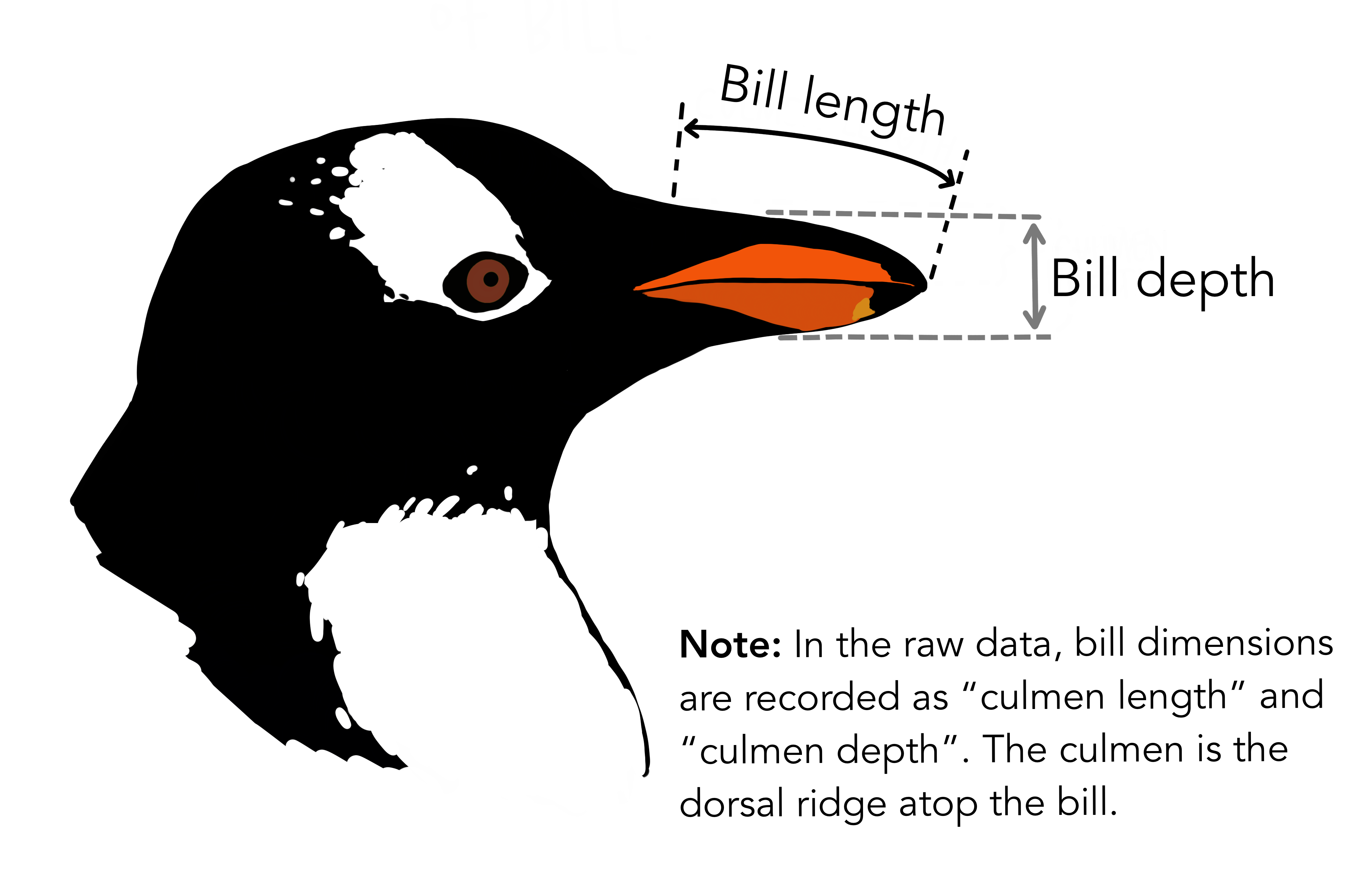
penguins dataset
Size measurements for adult foraging penguins near Palmer Station, Antarctica
Rows: 344Columns: 8$ species <fct> Adelie, Adelie, Adelie, Adelie, Adelie, Adelie, Ade…$ island <fct> Torgersen, Torgersen, Torgersen, Torgersen, Torgers…$ bill_length_mm <dbl> 39.1, 39.5, 40.3, NA, 36.7, 39.3, 38.9, 39.2, 34.1,…$ bill_depth_mm <dbl> 18.7, 17.4, 18.0, NA, 19.3, 20.6, 17.8, 19.6, 18.1,…$ flipper_length_mm <int> 181, 186, 195, NA, 193, 190, 181, 195, 193, 190, 18…$ body_mass_g <int> 3750, 3800, 3250, NA, 3450, 3650, 3625, 4675, 3475,…$ sex <fct> male, female, female, NA, female, male, female, mal…$ year <int> 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 200…
The Grammar of Graphics
Rows: 344Columns: 8$ species <fct> Adelie, Adelie, Adelie, Adelie, Adelie, Adelie, Ade…$ island <fct> Torgersen, Torgersen, Torgersen, Torgersen, Torgers…$ bill_length_mm <dbl> 39.1, 39.5, 40.3, NA, 36.7, 39.3, 38.9, 39.2, 34.1,…$ bill_depth_mm <dbl> 18.7, 17.4, 18.0, NA, 19.3, 20.6, 17.8, 19.6, 18.1,…$ flipper_length_mm <int> 181, 186, 195, NA, 193, 190, 181, 195, 193, 190, 18…$ body_mass_g <int> 3750, 3800, 3250, NA, 3450, 3650, 3625, 4675, 3475,…$ sex <fct> male, female, female, NA, female, male, female, mal…$ year <int> 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 200…
The Grammar of Graphics
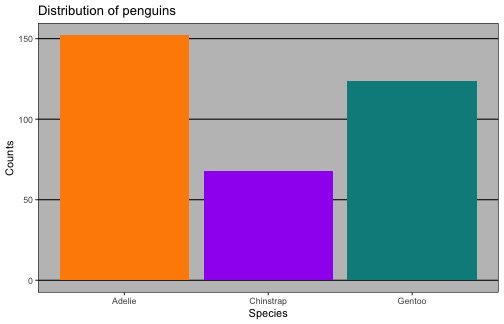
Rows: 344Columns: 8$ species <fct> Adelie, Adelie, Adelie, Adelie, Adelie, Adelie, Ade…$ island <fct> Torgersen, Torgersen, Torgersen, Torgersen, Torgers…$ bill_length_mm <dbl> 39.1, 39.5, 40.3, NA, 36.7, 39.3, 38.9, 39.2, 34.1,…$ bill_depth_mm <dbl> 18.7, 17.4, 18.0, NA, 19.3, 20.6, 17.8, 19.6, 18.1,…$ flipper_length_mm <int> 181, 186, 195, NA, 193, 190, 181, 195, 193, 190, 18…$ body_mass_g <int> 3750, 3800, 3250, NA, 3450, 3650, 3625, 4675, 3475,…$ sex <fct> male, female, female, NA, female, male, female, mal…$ year <int> 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 200…Counts
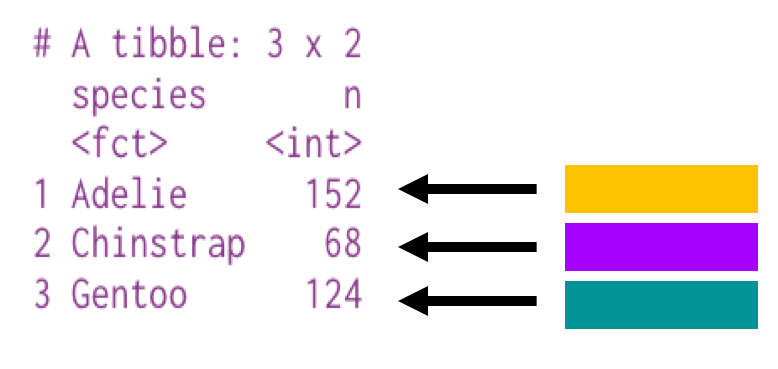
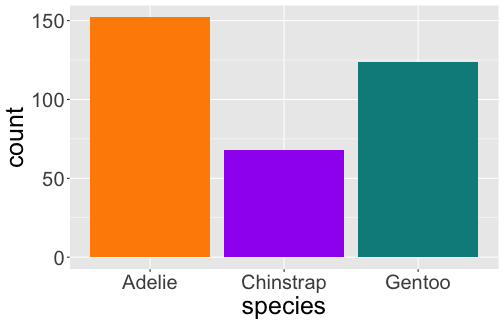
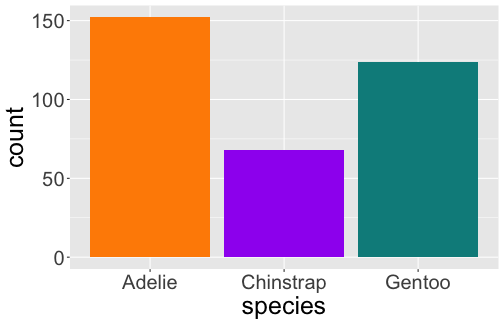
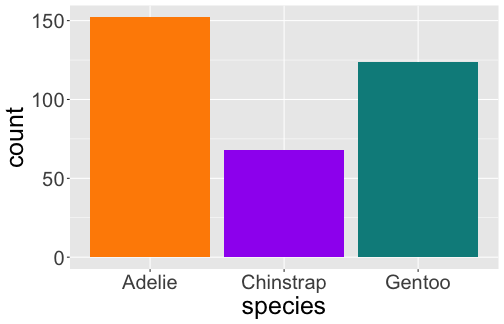
# A tibble: 3 x 2 species n <fct> <int>1 Adelie 1522 Chinstrap 683 Gentoo 124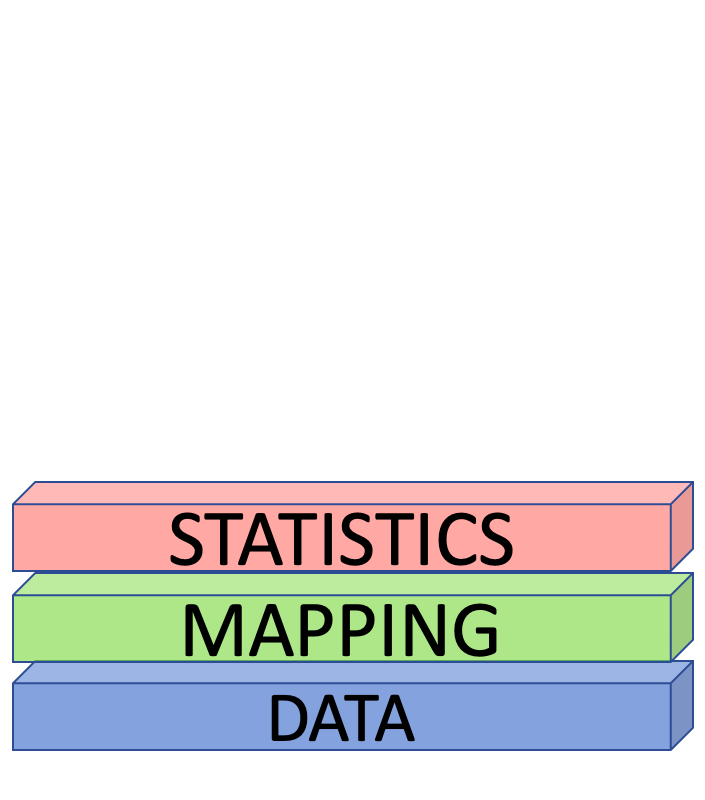
The Grammar of Graphics
The Grammar of Graphics

Bar Plot
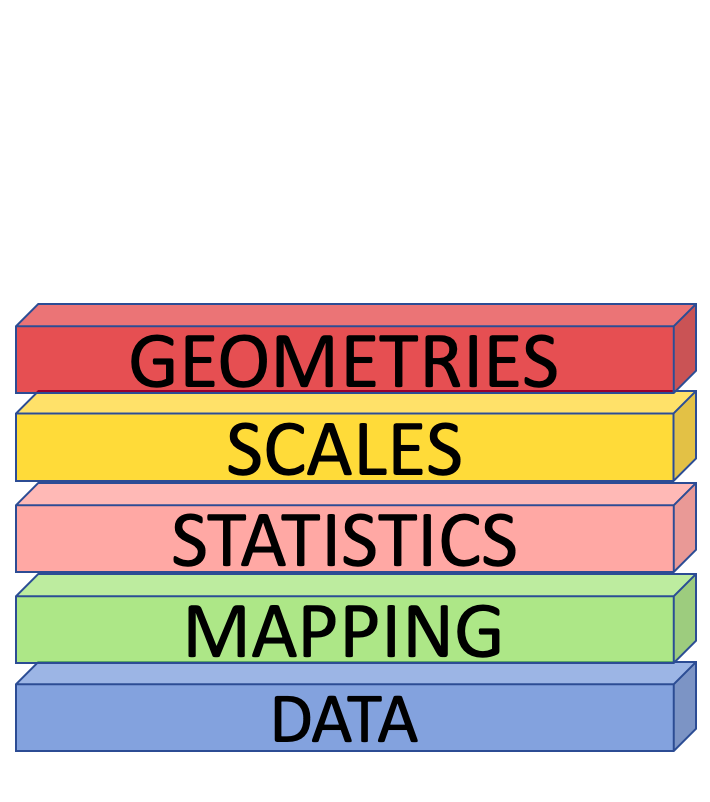
The Grammar of Graphics
Rows: 344Columns: 8$ species <fct> Adelie, Adelie, Adelie, Adelie, Adelie, Adelie, Ade…$ island <fct> Torgersen, Torgersen, Torgersen, Torgersen, Torgers…$ bill_length_mm <dbl> 39.1, 39.5, 40.3, NA, 36.7, 39.3, 38.9, 39.2, 34.1,…$ bill_depth_mm <dbl> 18.7, 17.4, 18.0, NA, 19.3, 20.6, 17.8, 19.6, 18.1,…$ flipper_length_mm <int> 181, 186, 195, NA, 193, 190, 181, 195, 193, 190, 18…$ body_mass_g <int> 3750, 3800, 3250, NA, 3450, 3650, 3625, 4675, 3475,…$ sex <fct> male, female, female, NA, female, male, female, mal…$ year <int> 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 200…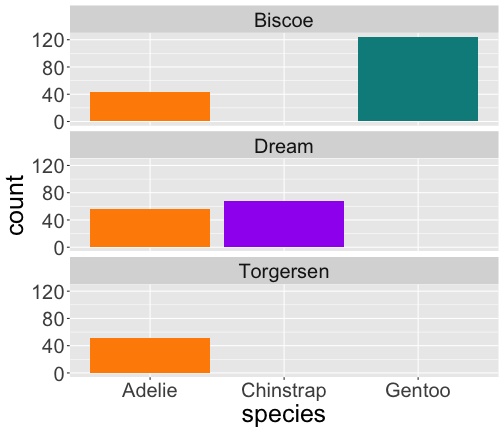
The Grammar of Graphics
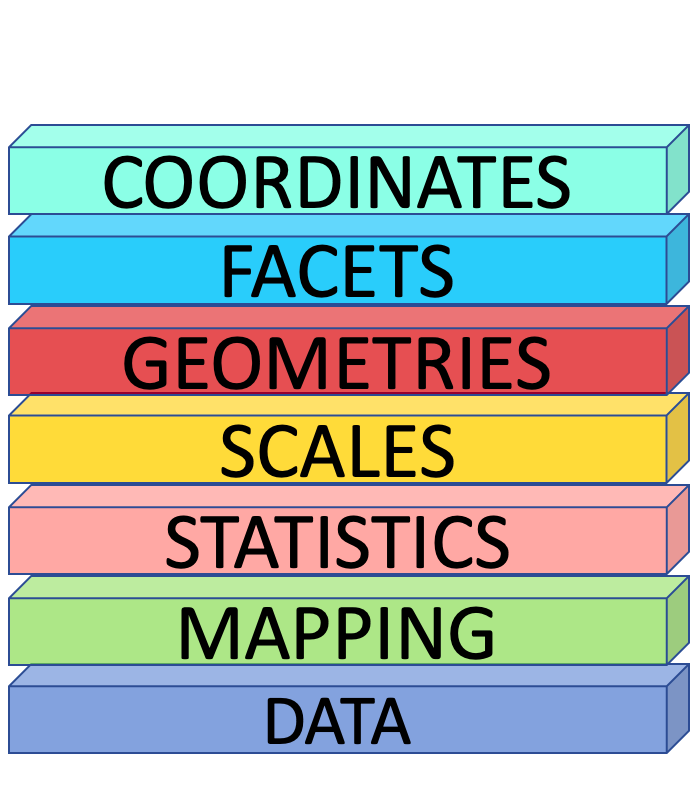
The Grammar of Graphics

The Grammar of Graphics
Excel Theme

The ggplot2 API

Which dataset to plot?

palmerpenguins data
The Palmer Archipelago penguins. Artwork by @allison_horst.


# A tibble: 6 x 8 species island bill_length_mm bill_depth_mm flipper_length_… body_mass_g sex <fct> <fct> <dbl> <dbl> <int> <int> <fct>1 Adelie Torge… 39.1 18.7 181 3750 male 2 Adelie Torge… 39.5 17.4 186 3800 fema…3 Adelie Torge… 40.3 18 195 3250 fema…4 Adelie Torge… NA NA NA NA <NA> 5 Adelie Torge… 36.7 19.3 193 3450 fema…6 Adelie Torge… 39.3 20.6 190 3650 male # … with 1 more variable: year <int>Rows: 344Columns: 8$ species <fct> Adelie, Adelie, Adelie, Adelie, Adelie, Adelie, Ade…$ island <fct> Torgersen, Torgersen, Torgersen, Torgersen, Torgers…$ bill_length_mm <dbl> 39.1, 39.5, 40.3, NA, 36.7, 39.3, 38.9, 39.2, 34.1,…$ bill_depth_mm <dbl> 18.7, 17.4, 18.0, NA, 19.3, 20.6, 17.8, 19.6, 18.1,…$ flipper_length_mm <int> 181, 186, 195, NA, 193, 190, 181, 195, 193, 190, 18…$ body_mass_g <int> 3750, 3800, 3250, NA, 3450, 3650, 3625, 4675, 3475,…$ sex <fct> male, female, female, NA, female, male, female, mal…$ year <int> 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 200…Which dataset to plot?
ggplot()
Which dataset to plot?
ggplot(data = penguins)
Mapping
Which columns to use for x and y?
ggplot(data = penguins, mapping = aes(x = flipper_length_mm, y = body_mass_g))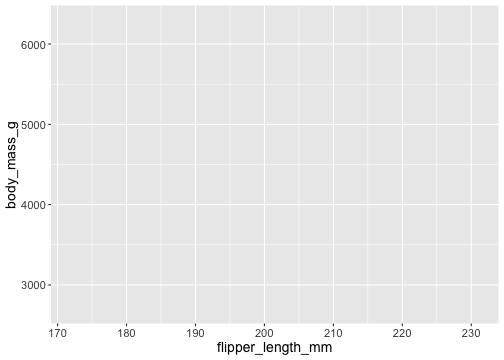
Geometries
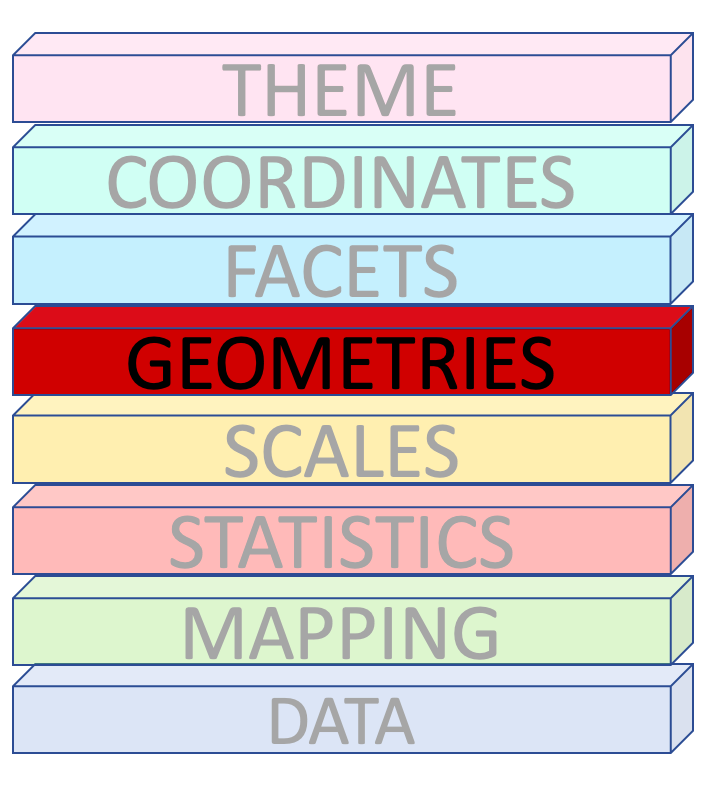
How to draw the plot?
ggplot(data = penguins, mapping = aes(x = flipper_length_mm, y = body_mass_g)) + geom_point()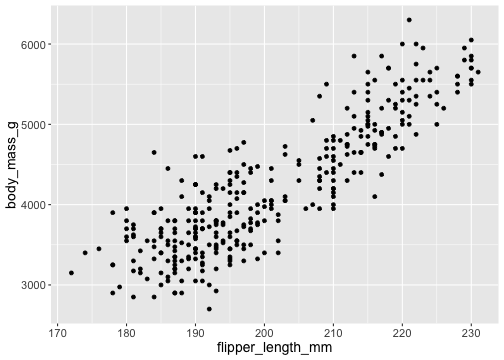
Data, Mapping and Geometries
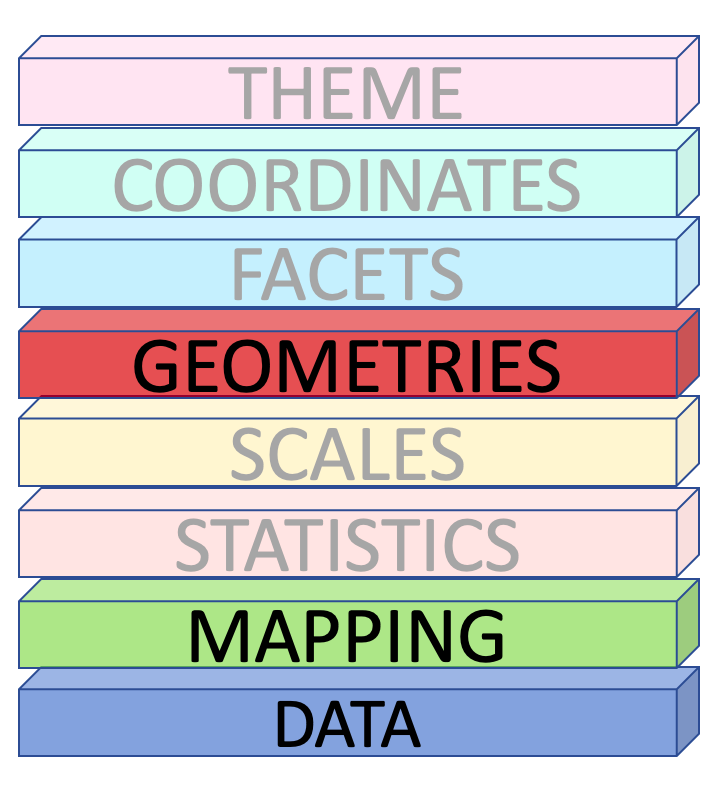
How to draw the plot?
ggplot(data = penguins) + geom_point(mapping = aes(x = flipper_length_mm, y = body_mass_g))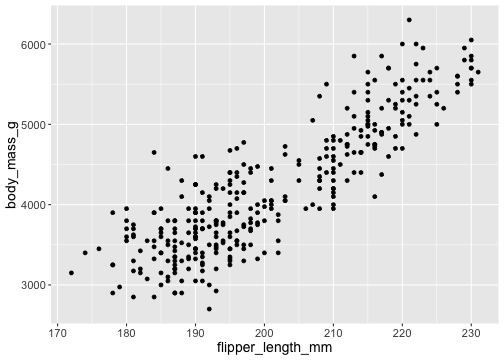
How to draw the plot?
ggplot() + geom_point(mapping = aes(x = flipper_length_mm, y = body_mass_g), data = penguins)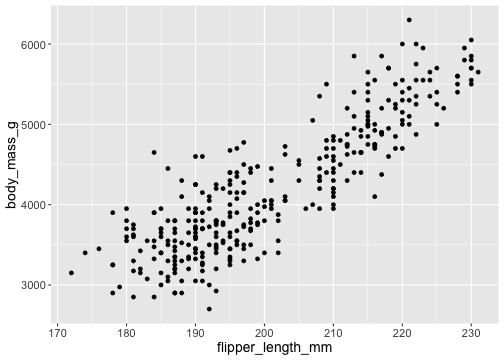
Mapping Colours
ggplot(penguins) + geom_point( aes(x = flipper_length_mm, y = body_mass_g, color = species, shape = species))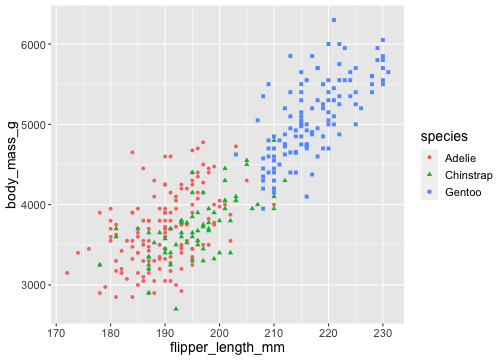
Mapping Colours
ggplot(penguins) + geom_point( aes(x = flipper_length_mm, y = body_mass_g, colour = flipper_length_mm < 205))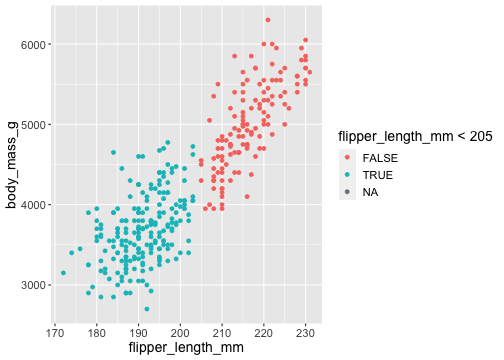
Setting Colours
ggplot(penguins) + geom_point( aes(x = flipper_length_mm, y = body_mass_g), colour = 'purple')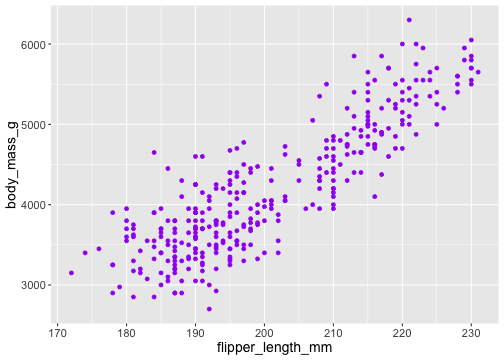
ggplot(penguins, aes(x = flipper_length_mm, y = body_mass_g, color = species, shape = species)) + geom_point() + geom_density_2d()- Syntax starts with
geom_*. - eg: geom_histogram(), geom_bar(), geom_boxplot().
- Each shape has its own specific aesthetics arguments.
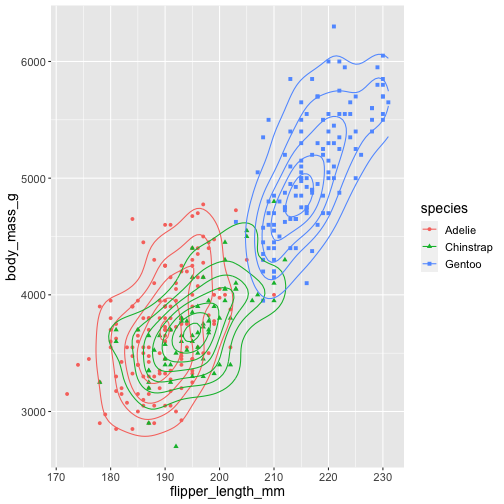
ggplot(penguins, aes(x = flipper_length_mm, y = body_mass_g, color = species, shape = species)) + geom_point() + geom_density_2d()- Syntax starts with
geom_*. - eg: geom_histogram(), geom_bar(), geom_boxplot().
- Each shape has its own specific aesthetics arguments.

ggplot(penguins) + geom_histogram( aes(x = flipper_length_mm))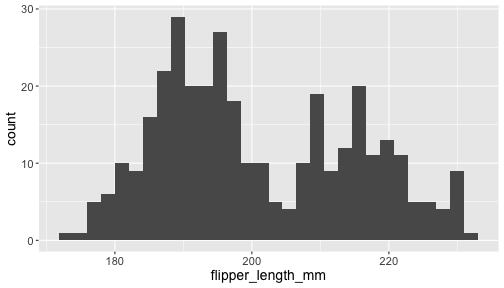
Each shape has its own specific aesthetics arguments.
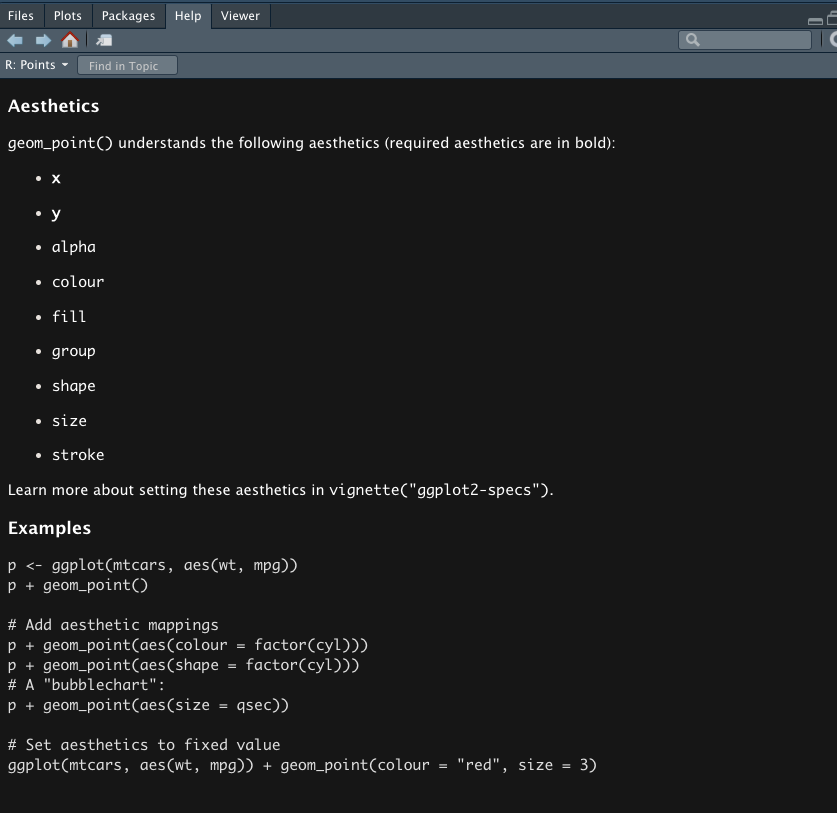
?geom_point
Statistics
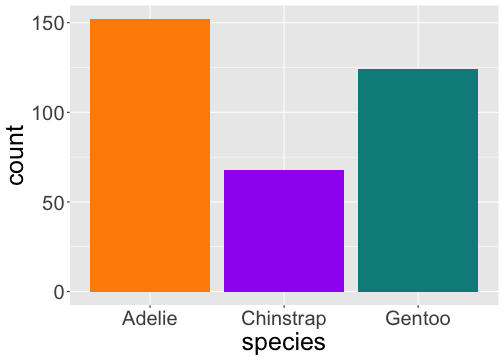
geom_bar() uses stat_count() by default
ggplot(penguins) + geom_bar(aes(x = species))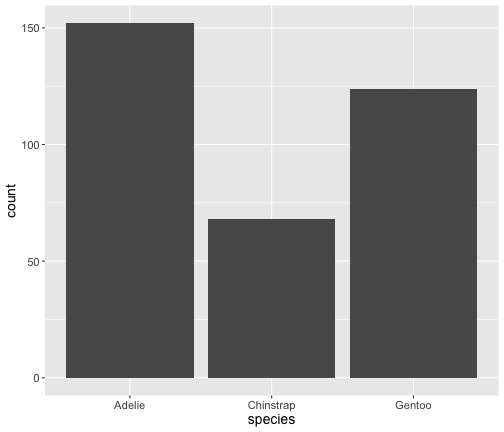
after_stat()
ggplot(penguins) + geom_bar(aes(x = species, y = after_stat(100*count/ sum (count)) ))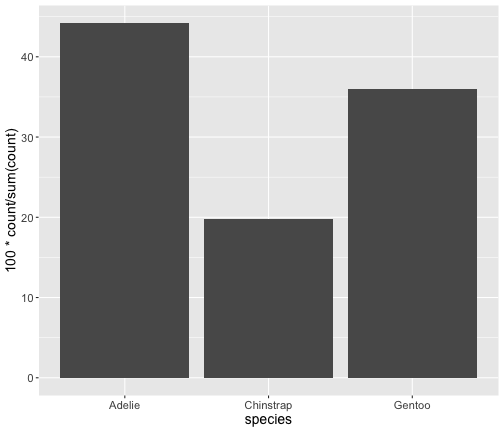
Old version of ggplot2
ggplot(penguins) + geom_bar(aes(x = species, y = 100*(..count..)/sum(..count..) ))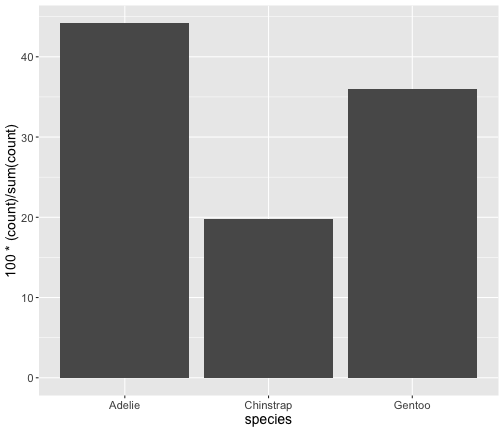
after_stat() (ggplot2 3.0.0)
ggplot(penguins) + geom_bar(aes(x = species, y = after_stat(100*count/ sum (count)) ))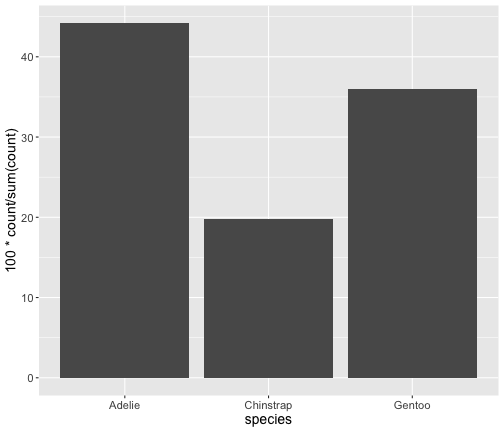
- There are two ways to use statistical functions.
define stat_*() function and geom argument inside that function
ggplot(penguins, aes(x = flipper_length_mm, y = body_mass_g)) + stat_summary( geom ="point", fun.y ="mean", colour ="red")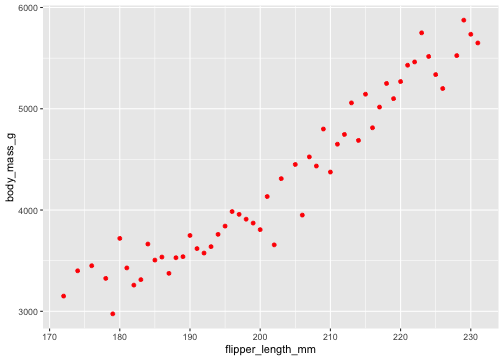
define geom_*() function and stat argument inside that function
ggplot(penguins, aes(x = flipper_length_mm, y = body_mass_g)) + geom_point( stat ="summary", fun.y ="mean", colour ="red")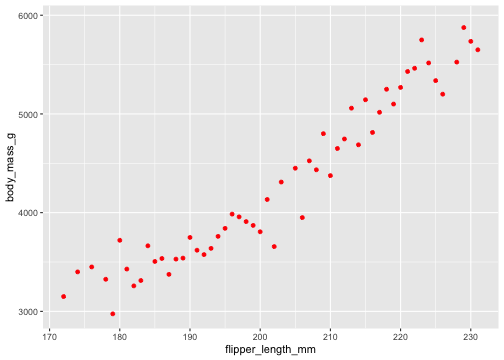
| Statistics | Geometries |
|---|---|
stat_count |
geom_bar |
stat_boxplot |
geom_boxplot |
stat_identity |
geom_col |
stat_bin |
geom_bar, geom_histogram |
stat_density |
geom_density |
| Statistics | Geometries |
|---|---|
stat_count |
geom_bar |
stat_boxplot |
geom_boxplot |
stat_identity |
geom_col |
stat_bin |
geom_bar, geom_histogram |
stat_density |
geom_density |
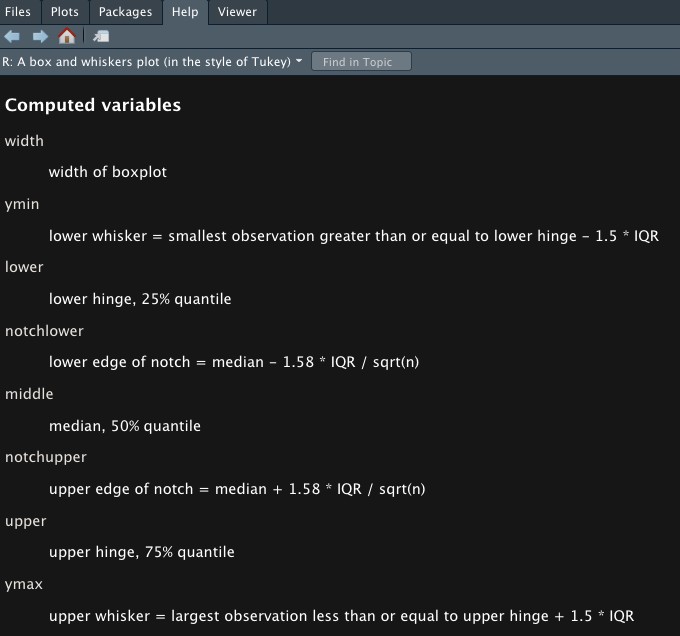
?geom_boxplot
?geom_bar
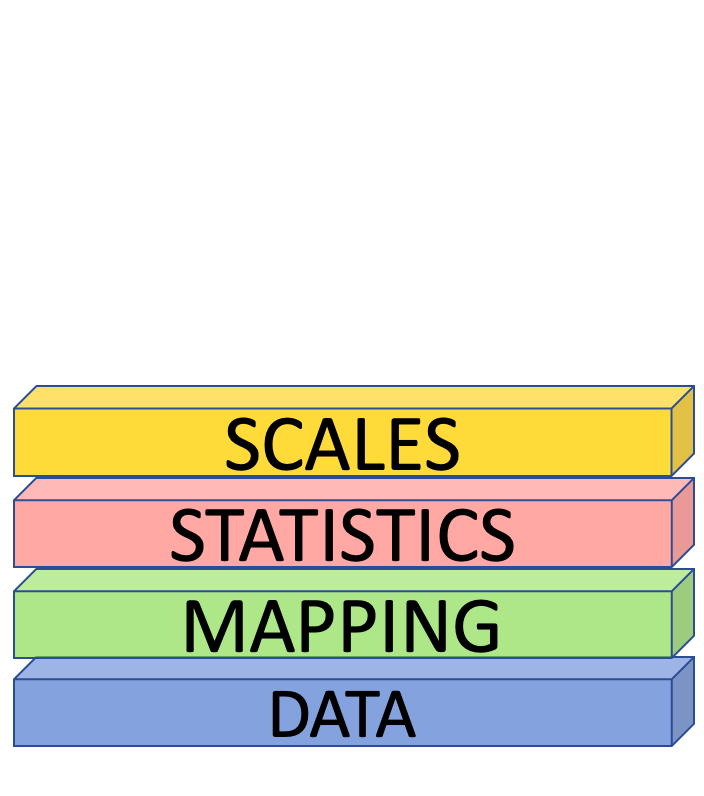
Scales
Scales
ggplot(penguins) + geom_point( aes(x = flipper_length_mm, y = body_mass_g, color = species, shape = species))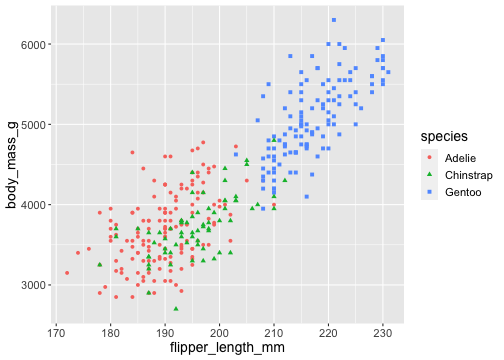
Scales
ggplot(penguins) + geom_point( aes(x = flipper_length_mm, y = body_mass_g, color = species, shape = island))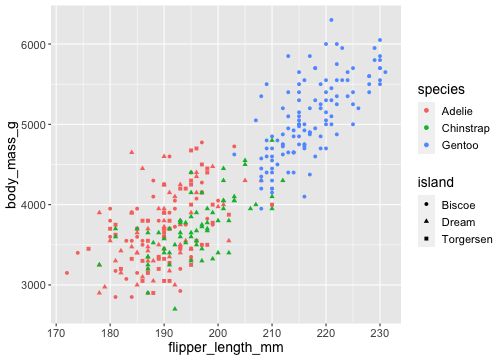
Scales
ggplot(penguins) + geom_point( aes(x = flipper_length_mm, y = body_mass_g, color = bill_length_mm, shape = island))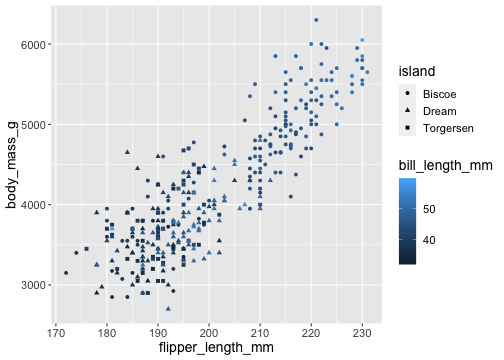
Scales
ggplot(penguins) + geom_point(aes(x = flipper_length_mm, y = body_mass_g, color = species)) + scale_color_brewer(type = 'qual', palette = 'Dark2')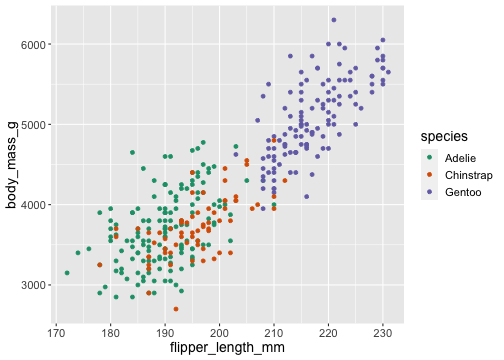
Scales
ggplot(penguins) + geom_point(aes(x = flipper_length_mm, y = body_mass_g, color = species)) + scale_color_brewer(type = 'qual', palette = 'Dark2')
scale_<aesthetic>_<type>
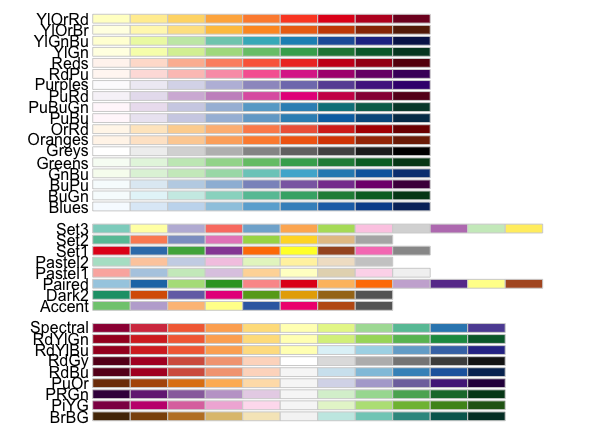
RColorBrewer::display.brewer.all()
ggplot(penguins) + geom_point(aes(x = flipper_length_mm, y = body_mass_g, color = species)) + scale_color_viridis_d()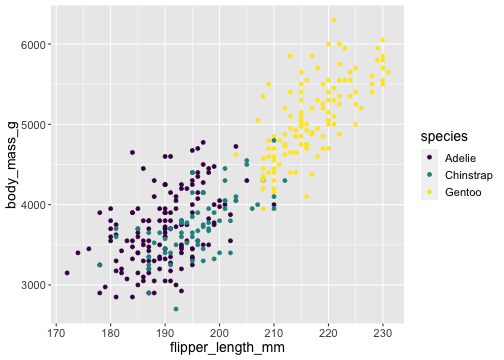
ggplot(penguins) + geom_point(aes(x = flipper_length_mm, y = body_mass_g, color = species)) + scale_color_viridis_d()
viridisandRColorBrewerprovide different color scales that are robust to color-blindness.
ggplot(penguins) + geom_point(aes(x = flipper_length_mm, y = body_mass_g, color = species)) + scale_color_viridis_d()
viridisandRColorBrewerprovide different color scales that are robust to color-blindness.- For details and an interactive palette selection tools see http://colorbrewer.org
ggplot(penguins) + geom_point(aes(x = flipper_length_mm, y = body_mass_g, color = species, shape = species, alpha = species)) + scale_x_continuous( breaks = c(170,200,230)) + scale_y_log10() + scale_colour_viridis_d(direction = -1, option= 'plasma') + scale_shape_manual( values = c(17,18,19)) + scale_alpha_manual( values = c( "Adelie" = 0.6, "Gentoo" = 0.5, # "Chinstrap" = 0.7))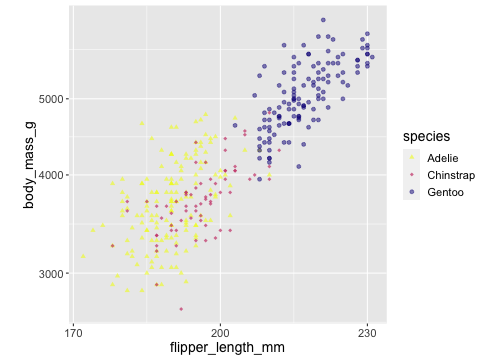
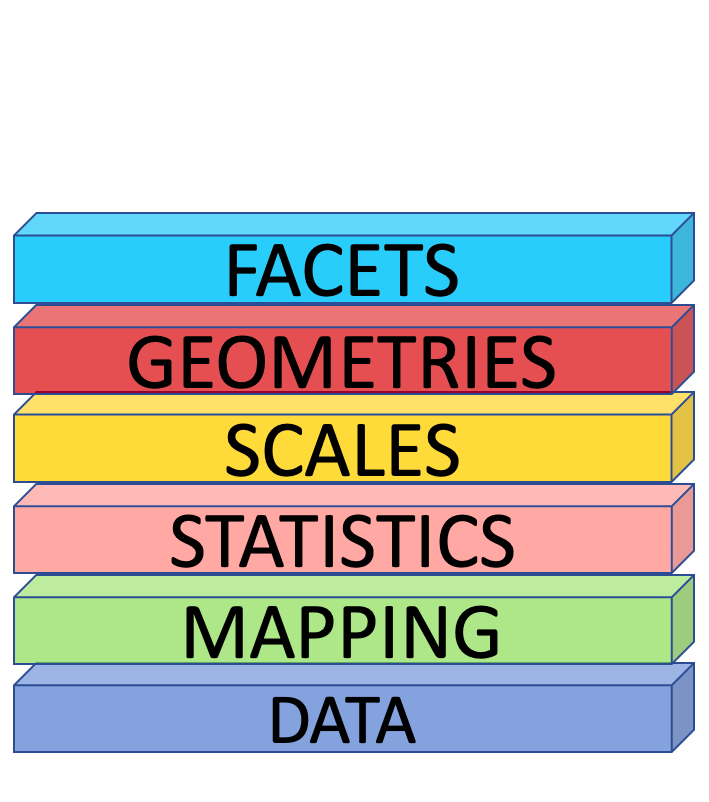
Facets
facet_wrap()
ggplot(penguins) + geom_point(aes( x = flipper_length_mm, y = body_mass_g)) + facet_wrap(vars(species))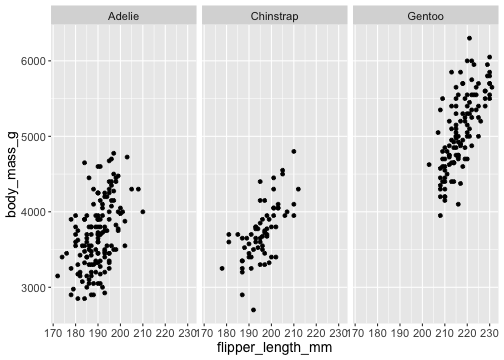
facet_wrap()
ggplot(penguins) + geom_point(aes( x = flipper_length_mm, y = body_mass_g)) + facet_wrap(vars(species), scales = "free_x")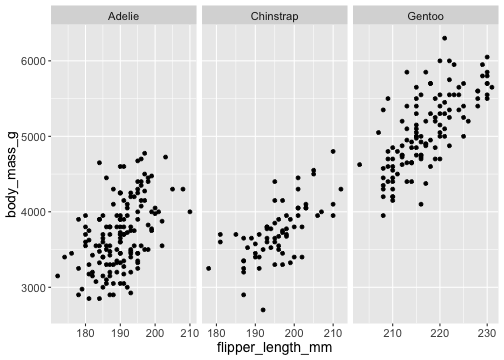
facet_grid()
ggplot(penguins) + geom_point(aes( x = flipper_length_mm, y = body_mass_g)) + facet_grid( vars(species), vars(sex))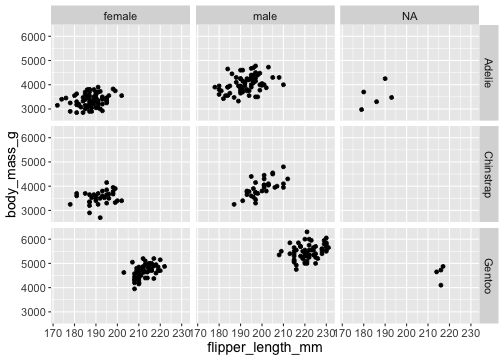
Coordinates
Coordinates
ggplot(penguins) + geom_bar(aes(x= species, fill = species))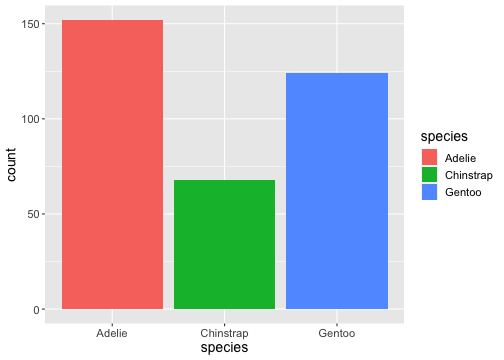
ggplot(penguins) + geom_bar(aes(x= species, fill = species)) + coord_flip()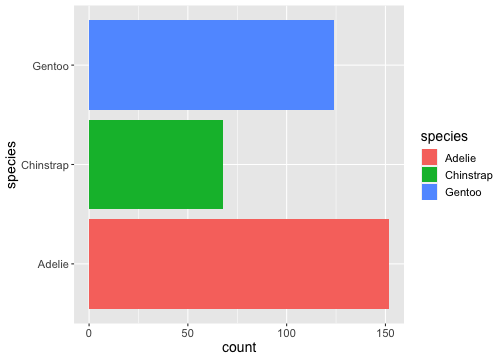
ggplot(penguins) + geom_bar(aes(x= species, fill = species)) + coord_flip()
- There are two types of coordinate systems:
- Linear coordinate systems
- Non-linear coordinate systems
ggplot(penguins) + geom_bar(aes(x= species, fill = species)) + coord_flip()
- There are two types of coordinate systems:
- Linear coordinate systems
- Non-linear coordinate systems
- Linear coordinate systems :
coord_cartesian(),coord_flip(),coord_fixed()
ggplot(penguins) + geom_bar(aes(x= species, fill = species)) + coord_flip()
- There are two types of coordinate systems:
- Linear coordinate systems
- Non-linear coordinate systems
- Linear coordinate systems :
coord_cartesian(),coord_flip(),coord_fixed() - Non-linear coordinate systems : eg :
coord_map(),coord_quickmap(),coord_sf(),coord_polar(),coord_trans()
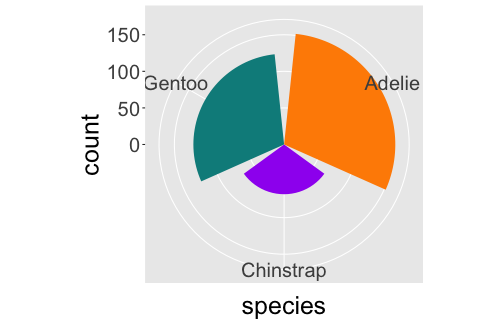
Coordinates
ggplot(penguins) + geom_bar(aes(x= species, fill = species)) + coord_polar()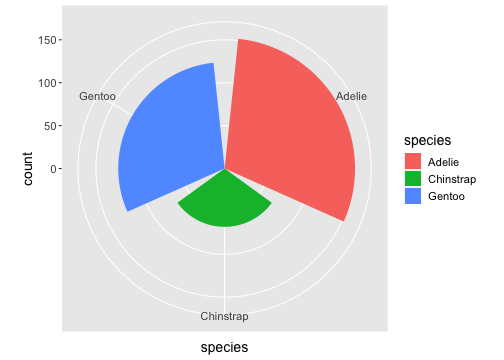
Coordinates
ggplot(penguins) + geom_bar(aes(x= species, fill = species)) + coord_polar(theta = "y")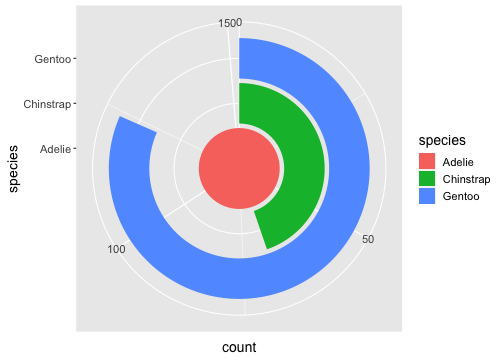
Zooming into a plot
ggplot(penguins) + geom_bar(aes(x= year, fill = species))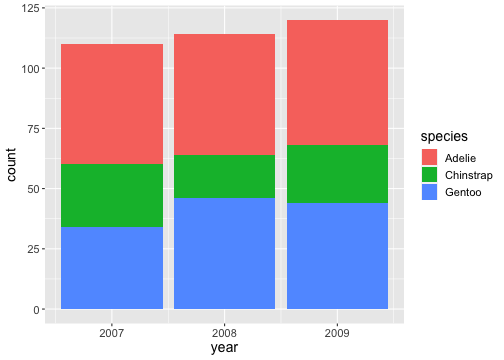
Zooming into a plot with scale
ggplot(penguins) + geom_bar(aes(x= year, fill = species)) + scale_y_continuous(limits = c(0,115))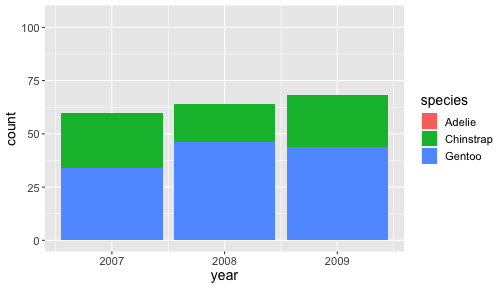
Zooming into a plot with scale
ggplot(penguins) + geom_bar(aes(x= year, fill = species)) + scale_y_continuous(limits = c(0,115))

When zooming with scale, any data outside the limits is thrown away
Proper zoom with coord_cartesian()
ggplot(penguins) + geom_bar(aes(x= year, fill = species)) + coord_cartesian(ylim = c(0,115))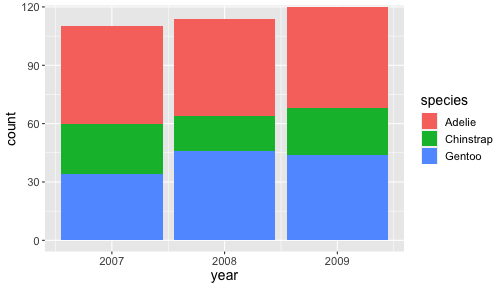
Proper zoom with coord_cartesian()
ggplot(penguins) + geom_bar(aes(x= year, fill = species)) + coord_cartesian(ylim = c(0,115))

Zooming with coord is like looking at the plot under a magnifying glass.
Themes
These are complete themes which control all non-data display.
ggplot(data = penguins, aes(x = flipper_length_mm, y = body_mass_g)) + geom_point(aes( color = species, shape = species), size = 3, alpha = 0.8) + theme_minimal()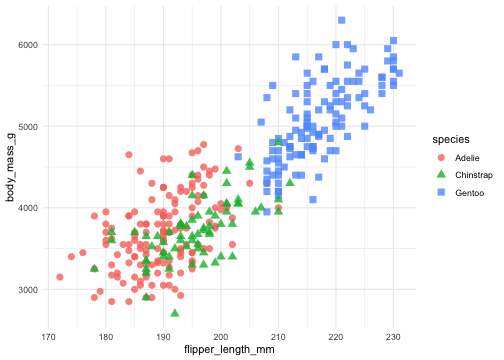
These are complete themes which control all non-data display.
ggplot(data = penguins, aes(x = flipper_length_mm, y = body_mass_g)) + geom_point(aes( color = species, shape = species), size = 3, alpha = 0.8) + theme_minimal()
ggplot(data = penguins, aes(x = flipper_length_mm, y = body_mass_g)) + geom_point(aes( color = species, shape = species), size = 3, alpha = 0.8) + theme_dark()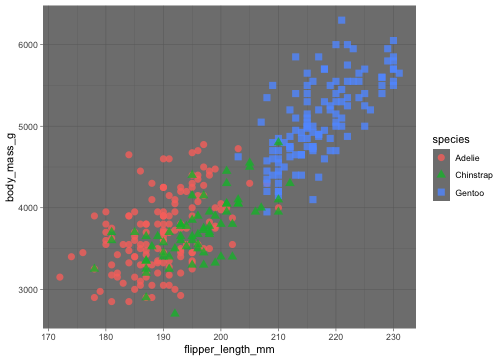
Create custom themes in ggplot.
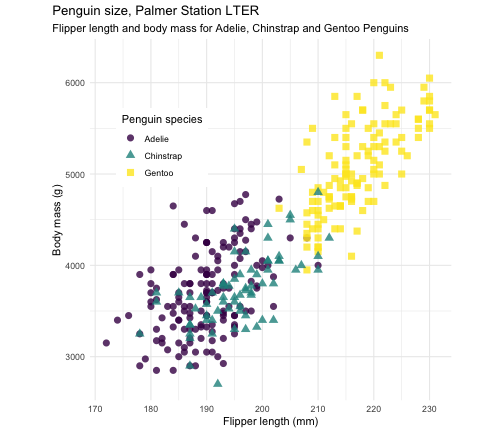
ggplot(penguins, aes(x = flipper_length_mm, y = body_mass_g)) + geom_point(aes(color = species, shape = species), size = 3, alpha = 0.8) + scale_color_viridis_d() + theme_minimal() + labs( title = "Penguin size, Palmer Station LTER", subtitle = "Flipper length and body mass for Adelie, Chinstrap and Gentoo Penguins", x = "Flipper length (mm)", y = "Body mass (g)", color = "Penguin species", shape = "Penguin species") + theme( aspect.ratio = 1, legend.position = c(0.2, 0.7), legend.background = element_rect( fill = "white", color = NA), plot.title.position = "plot", plot.caption = element_text( hjust = 0, face= "italic"), plot.caption.position = "plot")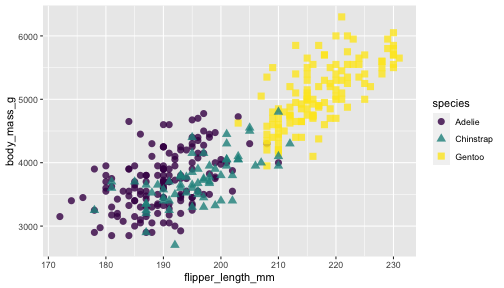
ggplot2 extensions
ggplot2 extensions: https://exts.ggplot2.tidyverse.org/
1. patchwork for plot composition

p1 <- ggplot(data = penguins, aes(x = flipper_length_mm, y = body_mass_g)) + geom_point(aes(color = species, shape = species), size = 2) + scale_color_manual(values = c("darkorange","darkorchid","cyan4")) + theme(aspect.ratio = 1)p2 <- ggplot(data = penguins, aes(x = bill_length_mm, y = bill_depth_mm)) + geom_point(aes(color = species, shape = species), size = 2) + scale_color_manual(values = c("darkorange","darkorchid","cyan4")) + theme(aspect.ratio = 1)p3 <- ggplot(data = penguins, aes(x = flipper_length_mm)) + geom_histogram(aes(fill = species), alpha = 0.5, position = "identity") + scale_fill_manual(values = c("darkorange","darkorchid","cyan4"))library(patchwork)p1 + p3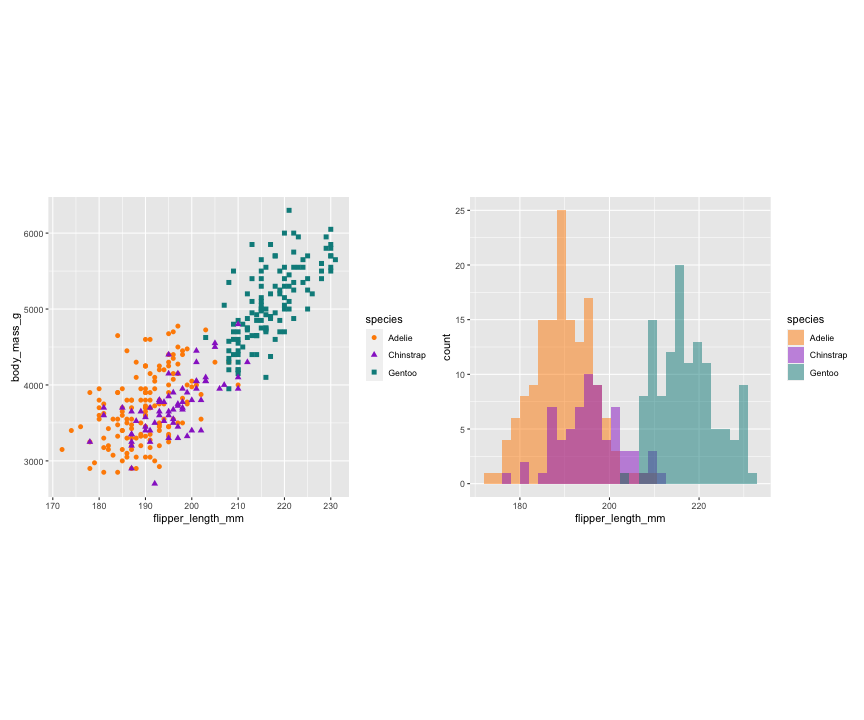
library(patchwork)(p1 | p2) / p3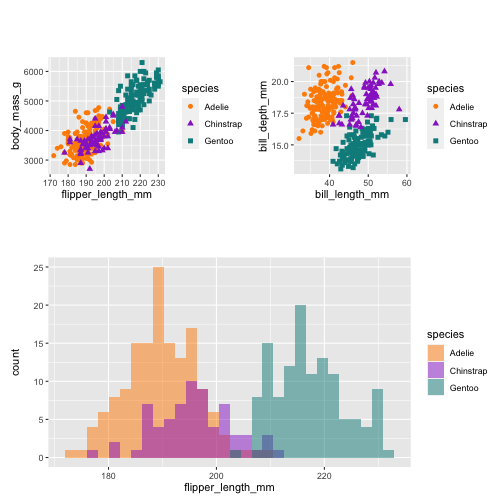
library(patchwork)p <- (p1 | p2) / p3p + plot_layout(guide = 'collect')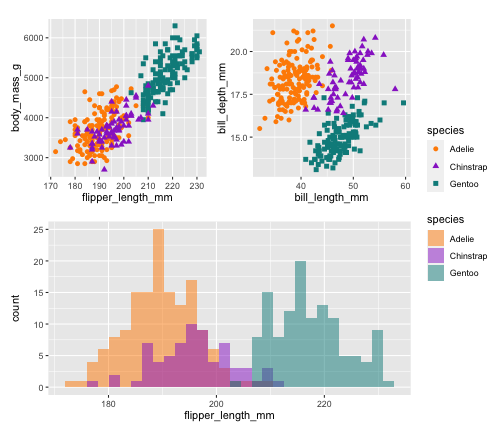
library(patchwork)p <- (p1 | p2) / p3p + plot_layout(guide = 'collect') + plot_annotation( title = 'Size measurements for adult foraging penguins near Palmer Station, Antarctica', tag_levels = 'A')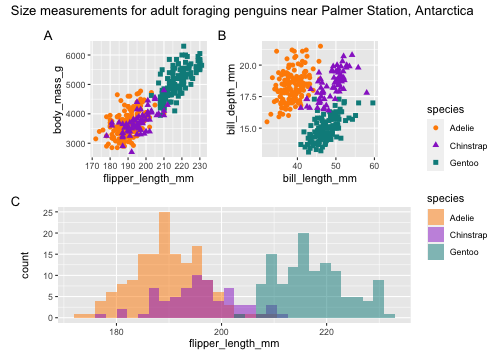
library(patchwork)p <- (p1 | p2) / p3p & theme(legend.position = 'none')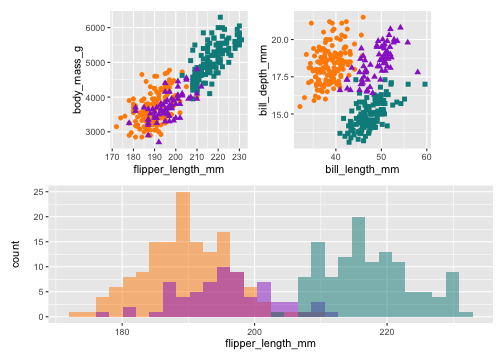
2. plotly
An R package for creating interactive web graphics via the open source JavaScript graphing library plotly.js.
p1 ## a ggplot object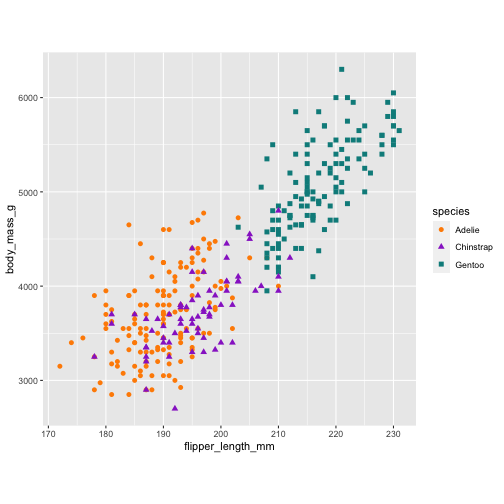
3. GGally
GGally::ggpairs(penguins[, 1:5], aes(color = species, fill = species))+ scale_color_viridis_d() + scale_fill_viridis_d()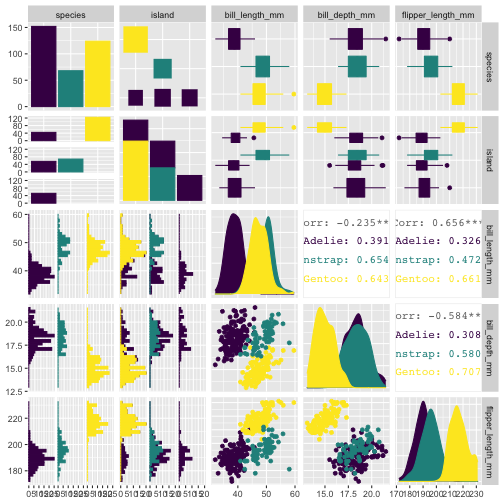
4. gganimate

library("ggplot2")library("dlstats")data <- cran_stats("ggplot2")p <- ggplot(data, aes(x= end, y = downloads)) + geom_line() + labs(title = "Download stats of ggplot2 package", x = "Time", y = "Downloads")p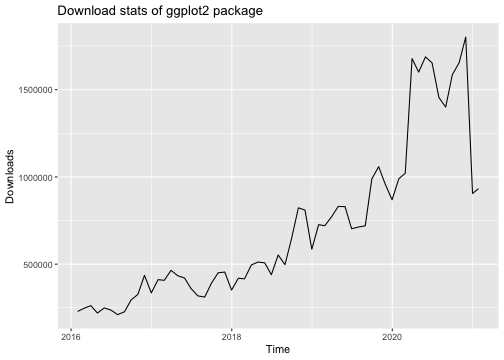
library(gganimate)p + transition_reveal(along = end)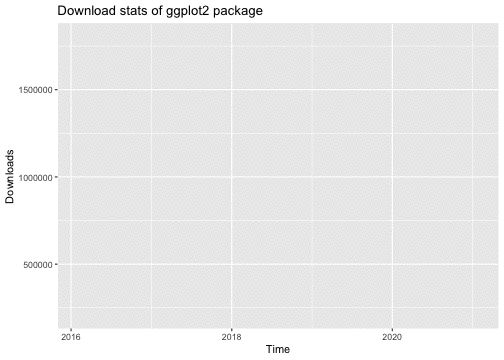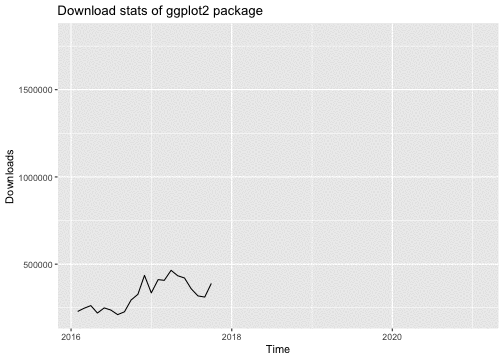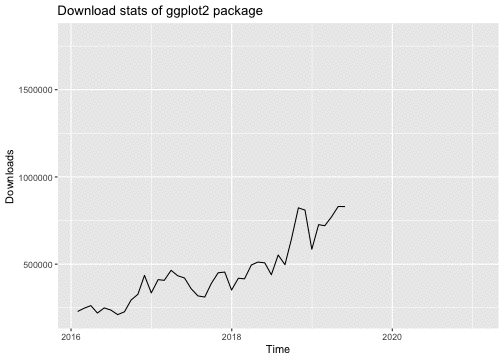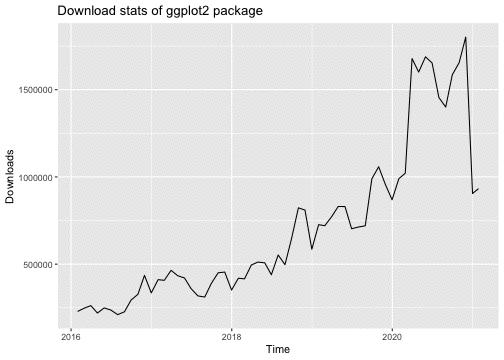
p <- ggplot(penguins, aes(flipper_length_mm, body_mass_g , color = species)) + geom_point() + scale_color_viridis_d() + labs(title = "Measurements of penguins {closest_state}")+ transition_states(states = year) + enter_grow() + exit_fade()p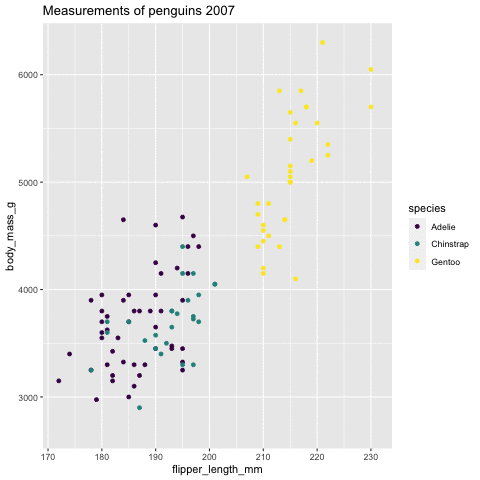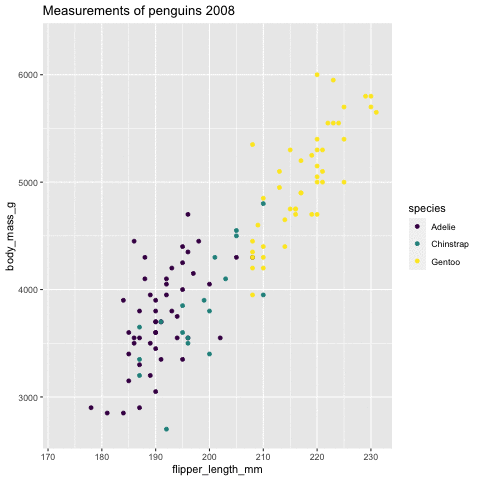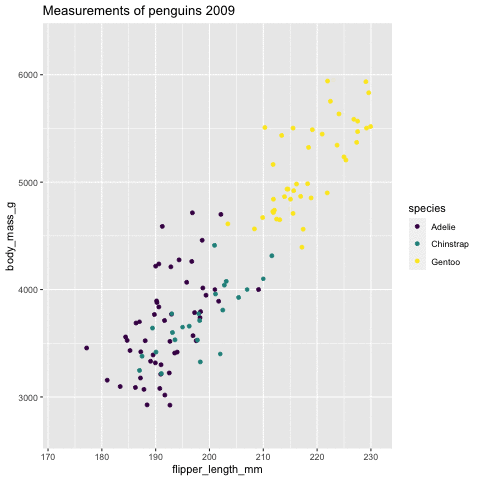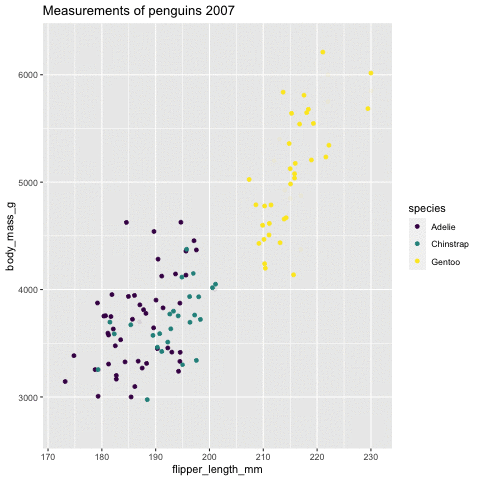
5. ggrepel

Text annotation
df <- penguins %>% filter( flipper_length_mm > 225 )ggplot(penguins, aes(x=flipper_length_mm, y= body_mass_g))+ geom_point()+ theme(aspect.ratio = 1) + geom_text(data= df, aes(x=flipper_length_mm, y= body_mass_g, label= island))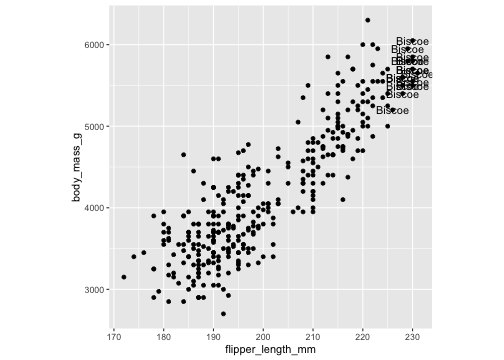
Text annotation
ggplot(penguins, aes(x=flipper_length_mm, y= body_mass_g))+ geom_point()+ theme(aspect.ratio = 1) + ggrepel::geom_text_repel(data= df, aes(x=flipper_length_mm, y= body_mass_g, label= island))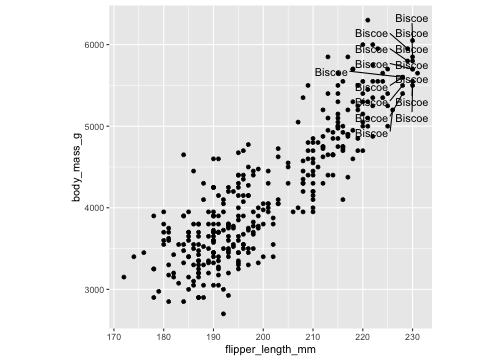
6. ggforce

library(ggforce)penguins <- penguins %>% drop_na()p <- ggplot(penguins, aes(x=flipper_length_mm, y= body_mass_g))+ geom_mark_ellipse(aes( filter = species == "Gentoo", label = 'Gentoo penguins'), description = 'Palmer Station Antarctica LTER and K. Gorman. 2020.') + geom_point() p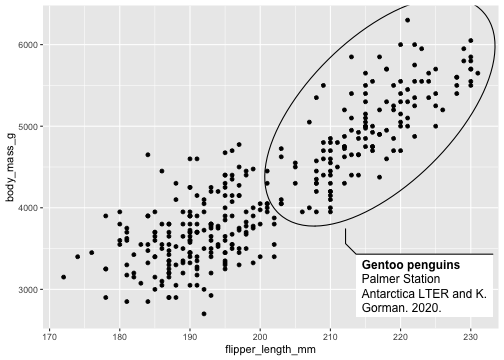
library(ggforce)ggplot(penguins, aes(x=flipper_length_mm, y= body_mass_g, color = species)) + geom_point() + scale_color_viridis_d() + facet_zoom(x = species == "Gentoo")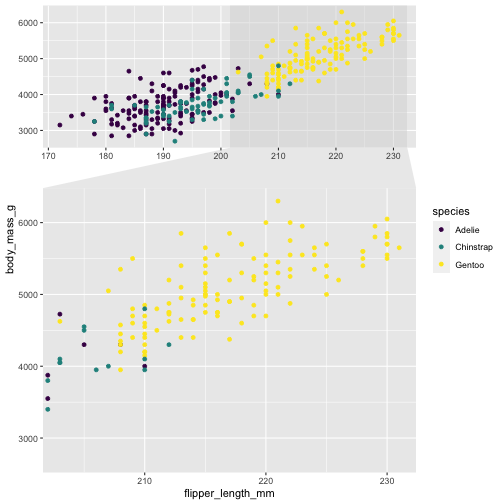
Thank you
Slides available at: prital.netlify.app
pridiltal
Acknowledgements:
Hadley Wickham, Thomas Lin Pedersen and ggplot2 development team
Key References
- ggplot2: Elegant Graphics for Data Analysis https://ggplot2-book.org/
- ggplot2 workshop by Thomas Lin Pedersen https://www.youtube.com/watch?v=h29g21z0a68
This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.